A new look inside Ebola’s “viral factories”
By Newswise
La Jolla, CA–New research in the journal Nature Communications gives scientists an important window into how Ebola virus replicates inside host cells.
The study, led by scientists at La Jolla Institute for Immunology (LJI), reveals the inner workings of “viral factories,” clusters of viral proteins and genomes that form in host cells.
The research team, which included experts from Scripps Research and UC San Diego School of Medicine, found that Ebola virus’s replication machinery forms fascinating microscopic structures that become viral factories. By understanding the architecture and function of these microscopic manufacturing hubs, researchers may be closer to developing new therapies that interrupt the Ebola virus life cycle and prevent severe disease.
“We are imaging these fluid and dynamic assembly centers for the first time. Understanding how they work and what they require gives us the information needed to defeat them,” says LJI President and CEO Erica Ollmann Saphire, Ph.D., senior author of the new study.
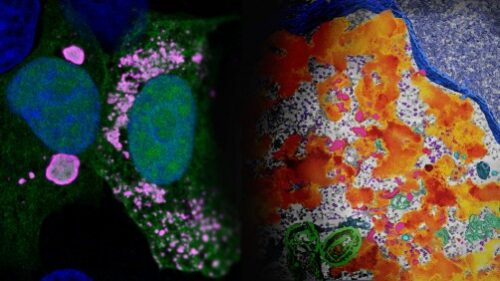
Scientists first spotted what would turn out to be “virus factories” in virus-infected animal cells back in the 1960s, but they didn’t know what they were seeing. Within a sea of normal cellular proteins, these areas looked like fuzzy splotches.
“People had already seen that Ebola-infected cells had these ‘inclusions,’” says LJI Postdoctoral Researcher Jingru Fang, Ph.D., first author of the new study. For a long time, scientists thought of these “inclusions” as helpful visual indicators of infection, without understanding their true purpose. “But in fact, these ‘inclusion bodies’ actively gather an enormous quantity of viral proteins and viral RNAs.”
Many viral pathogens, including rabies virus and RSV (respiratory syncytial virus) form inclusions in host cells, Fang explains. “Recent studies suggest that these cellular inclusions are the site where viruses make their RNA genomes. They are ‘viral factories’ with actual functional purpose: to offer a secured space for viral RNA synthesis,” says Fang. “The process of viral RNA synthesis involves flux of viral building blocks. This means molecules gathered inside viral factories should be able to move freely rather than being static.”
For the new study, Saphire, Fang, and their colleagues wondered: Can we observe the movement of viral building blocks directly in living cells?
Fang began by tagging a viral protein called VP35 with a fluorescent marker that makes the protein glow in the dark. VP35 is a critical component of the viral factory and is important for viral RNA synthesis (and the making of new copies of Ebola virus). Working with imaging experts in the LJI Microscopy and Histology Core, Fang followed the glowing proteins in live cells, which express a simplified and non-infectious version of Ebola viral factories.
Under the microscope, Fang and colleagues could indeed see and even measure how molecules move inside the viral factories formed in host cells. This finding added to evidence that viral proteins are clumping together like droplets so they can churn out the proteins needed to help the virus replicate. Those mysterious inclusions really are viral factories. The researcher dubbed these “droplet-like” viral factories.
Then the scientists saw something odd. Some of the glowing proteins didn’t gather into clumps. Instead, they joined up with a smattering of other viral proteins, creating a fluorescent swirl that evoked van Gogh’s “Starry Night.” These trails of viral proteins still had the right ingredients to replicate Ebola virus, so the scientists dubbed them “network-like” viral factories.
“These are two different flavors of the viral factory,” says Fang. “People have mostly focused on the droplet-like form, which is the majority, and not paid too much attention to this other form.”
Besides their shapes, there was a key difference between the two factories. It appeared the network-like factories had the right ingredients for the incoming Ebola virus to express its genes, but they didn’t actually produce virus progenies.
Next, the researchers looked at a key player in infection: a protein called virus polymerase. Polymerase is a multifunctional nanomachine that comes with the virus. This machine not only copies the Ebola virus genomic material, it also transcribes the viral genome into messenger RNAs, which instruct infected cells to produce loads of viral proteins. The researchers wanted to understand how this viral machine functions inside viral factories.
Ebola virus polymerase is already known as a hard-working protein—all Ebola viral proteins have to be. Ebola virus is a highly efficient pathogen because it gets by with just seven genes (humans have more than 20,000 genes). Saphire has led research showing that Ebola virus survives by making proteins that can transform and take on different jobs during the course of infection.
Just last year, Saphire, Fang, and collaborators published a related discovery that viral polymerase actually harnesses a druggable human protein to help the virus replicate its genome. The team reported that while polymerase is essential for viral replication, the polymerase doesn’t actually jump into action until the infection is well underway.
This work was important for understanding how polymerase stepped into action, but scientists also needed to know where polymerase was active. Fang knew it would be important to look at what polymerase might be up to in viral factories.
The researchers discovered that polymerase actually builds its own special structures inside viral factories. Many copies of polymerase gather in small bundles, called foci. The researchers found that these bundles spread out when a droplet-like viral factory starts replicating viral material.
Scientists aren’t sure exactly why polymerase needs to form bundles before it can do its job, but the spatial arrangement of the bundles must be important. As Fang points out, the idea of many small components coming together to build a structure isn’t a new concept in nature. “You can use a beehive or coral reef as the analogy to help understand why a specific spatial arrangement is important for a biological system to function,” she says.
With this finding, scientists now know how to find different kinds of viral factories and how polymerase organizes itself down on the factory floor.
More than 30 human pathogens are known to assemble viral factories inside host cells, including respiratory syncytial virus (RSV) and even rabies virus. With this new view of Ebola’s viral factories, scientists are curious whether other viruses construct similar forms of viral factories—and whether other viruses use their own versions of polymerase in the same way.
“If that’s true, maybe we can target the feature of viral factory formation that has been shared by multiple different viruses,” says Fang.
Going forward, Fang would also like to study how Ebola virus forms viral factories in different kinds of host cells. Do these viral factories look different in cells from animals (such as the virus’s natural hosts, the fruit bats) that can carry the virus around without getting sick? “Can we find some explanation for host-specific viral pathogenesis?” she asks.
The new study also demonstrates the importance of collaboration across San Diego’s Torrey Pines Mesa. The LJI team worked closely with Scripps Research Professor Ashok Deniz, Ph.D., and UC San Diego Professor Mark H. Ellisman, Ph.D., Director of the National Center for Microscopy and Imaging Research.
“The combination of state-of-the-art tools available on the Torrey Pines Mesa allowed us to combine the biophysical characterization with the human health insight,” says Saphire
Additional authors of the study, “Spatial and functional arrangement of Ebola virus polymerase inside phase-separated viral factories,” include Guillaume Castillon, Sebastien Phan, Sara McArdle, Chitra Hariharan, and Aiyana Adams.
This study was supported by the National Institute of Health, an Imaging Scientist grant from the Chan Zuckerberg Initiative, LJI institutional funds, and the Donald E. and Delia B. Baxter Foundation Fellowship.





